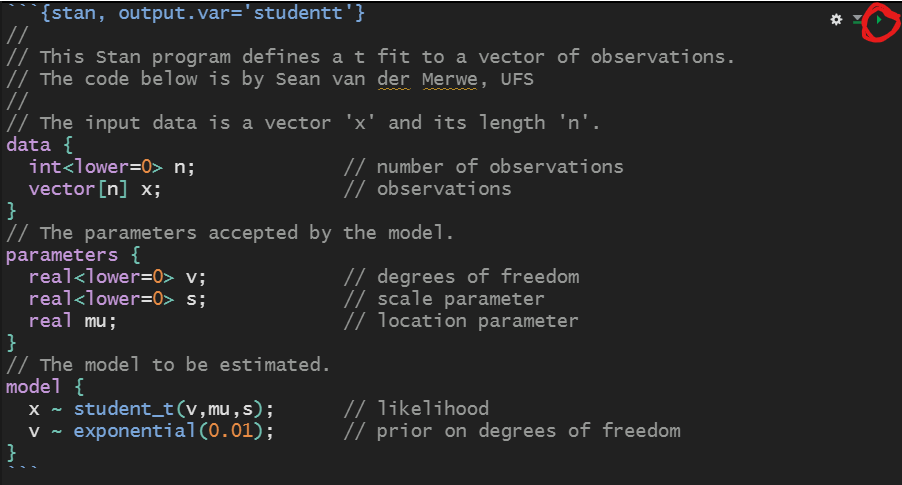
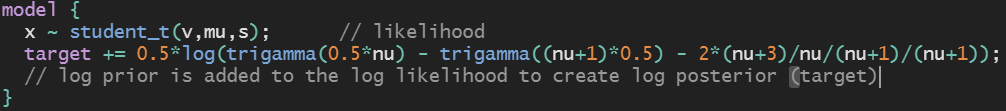
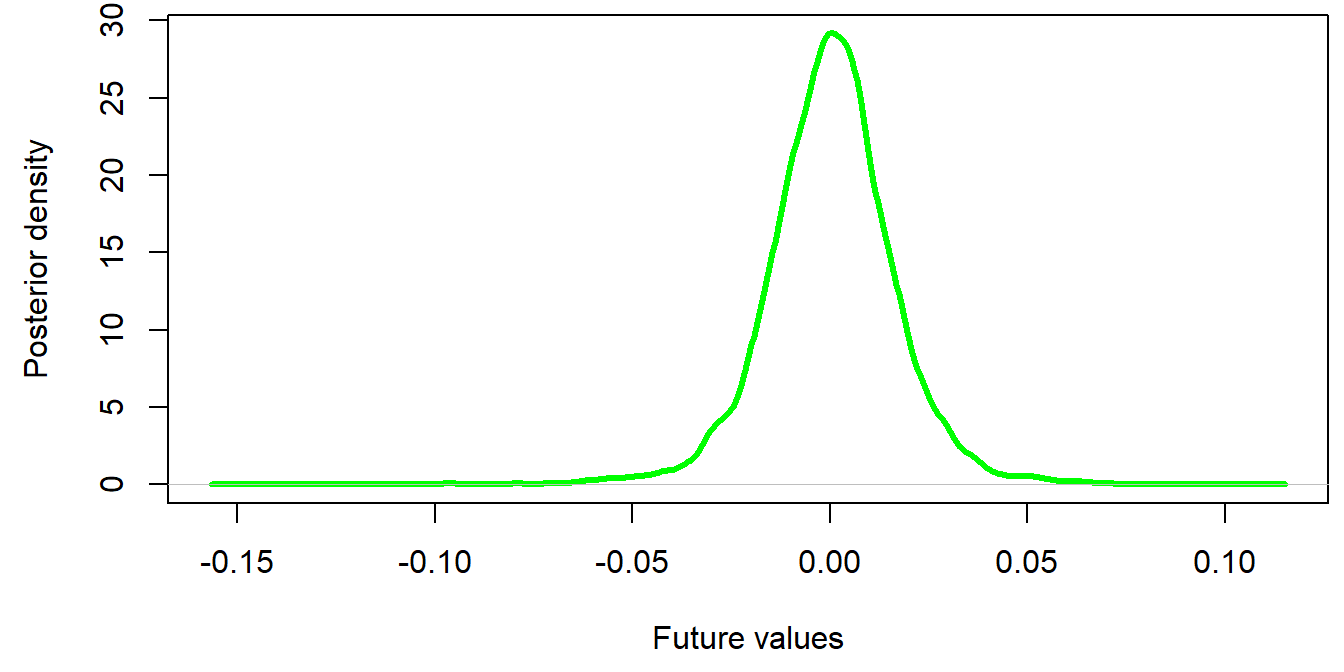
Please open the presentation on your own computer (type link above) to enjoy the following benefits:
Exploring the data yourself
Interacting with the graphs yourself
Moving at your own pace (press o for overview)
Maximum quality
- Try f or F11 for full screen
- Press w at any time to change to wide screen mode and back
- Ctrl + Mouse Wheel to zoom
The presentation and source code will be available for download on my website at seanvdm.co.za/files/workshops/mdag2021